Coaggregation: Interactions between Bacteria in Dental Biofilms
Contributed by:
- Daniel L. Clemans and Lindsey R. Kolar (Department of Biology, Eastern Michigan University)
- John Lennox (Department of Microbiology, Penn State Altoona) Emeritus
Instructor Version (go to Student Version)
| Subject Area(s) | Microbiology, Allied Health, Microbial Ecology |
Intended Audience |
High school biology, independent study/science fair, introductory microbiology, advanced college microbiology |
Type |
Laboratory exercise |
Revision Date |
July 12, 2010 |
CONTENT
In 1970, Gibbons and Nygaard observed that when pure cultures of some dental plaque bacterial strains were mixed, the suspension rapidly cleared. For example, when broth cultures of Actinomyces naeslundii and certain strains of Streptococcus sanquinis were combined a marked decrease in turbidity occurred within minutes1. This phenomenon, now termed coaggregation, is of selective value to bacterial living in a flowing environment as any cells, which detach from the oral surface are washed away and swallowed. Kolenbrander and others working on this phenomenon have found that coaggregation is exhibited by nearly all oral bacteria tested, a sample which includes more than seven hundred bacterial strains representing at least 18 genera2,3,4,5.
Figure 1 shows a diagrammatic sketch of the arrangement of bacterial strains which form dental biofilms. An acquired pellicle or conditioning film composed of proteins drawn from the saliva or crevicular fluid first attaches to the tooth surface. Pioneering bacteria, largely streptococci, attach to these proteins. Other organisms attach to these pioneers largely by means of coaggregation between a protein lectin on one organisms and a carbohydrate receptor on the other From Rickert et al. 2003, used with permission.
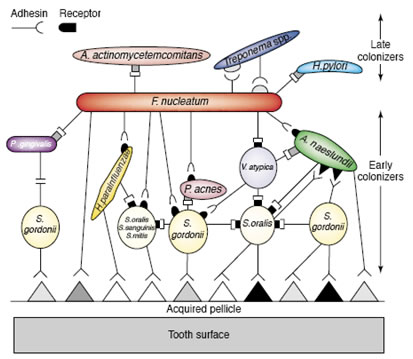
The order in which bacteria colonize the surface of teeth is, in large part, determined by the availability of bacterial species and the coaggregation pairings between them. In many instances, the mere availability of a particular strain is insufficient to guarantee its inclusion in the dental biofilm unless specific coaggregation partners have attached previously. Thus, biofilm formation is not random, but follows a specific pattern of succession. It remains to be seen if this phenomenon is entirely general or found in only certain environments.
Early colonizers, predominantly Streptococci attach to a conditioning film composed of proteins derived from the saliva or crevicular fluid. Subsequently, actinomycetes and Fusobacteria bind by coaggregation to these pioneer species. Depending largely upon the effectiveness of dental hygiene, other organisms join the biofilm. These late arriving species have little ability to coaggregate with the pioneers, but do bind with the intermediates such as Fusobacterium. The fusobacteria are therefore called bridging species (Figure 1).
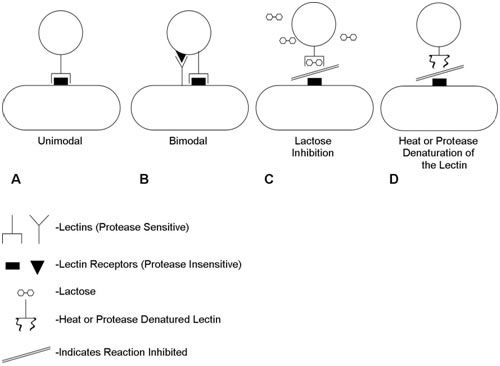
It is now known that the dominant cell-cell interactions are between lectin type (protein) adhesins on one of the cells and oligosaccharide moieties on the other. Coaggregation can be interrupted by denaturation of the lectin or, frequently, by the addition of sugar (e.g. lactose) which blocks the active lectin site by competitive inhibition (Figure 2). Figure 2 shows the nature of the interactions between coaggregating pairs of organisms. Some pairs have just a single known lectin/receptor pair (unimodal) while others may have two (bimodal). Coaggregation may be inhibited either by denaturation of the protein lectin or by competitive inhibition of the lactin/cagbohydrate interaction by a sugar (e.g. lactose).
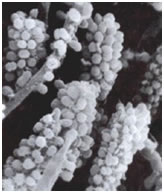
Among the most interesting morphological structures observable in samples of oral biofilms are the so-called “corncobs” or “test tube brush formations”, by the investigators who first described them (Figure 3: Corncob formations consisting of cocci coaggregating with a filamentous bacterium, possibly streptococcus and a fusobacterium. Used with permission of SaneDentist - http://www.sanedentist.com).
In these structures cocci coaggregate with rod-shaped or filamentous bacteria producing the characteristic corncob shape. These can occasionally be seen in negative stained preparations of dental tartar or plaque viewed under the brightfield microscope. An analogous formation called a “rosette” consists of a single cell of one bacterial strain coaggregating with a larger number of cocci of a different type completely surrounding the first.
Dental biofilms “grow” by two mechanisms, accretion in which additional microorganisms are added successively to the community, increasing its diversity and cell division of the individual cells forming microcolonial islands which contribute to the biofilms mass (Figure 4).
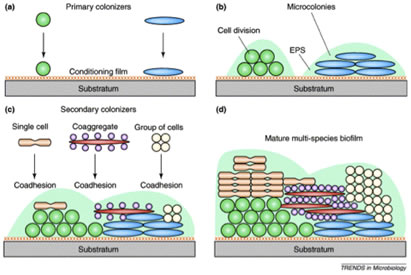

Specific adhesins on one member of a pair of organisms may bind to cognate receptors on the other forming masses of bacteria the size of which depends on the strength of the coaggregation reaction. These masses form very rapidly and can be easily viewed with the naked eye, although back-lighting and some magnification makes quantification much easier. All three of the experiments described are carried with materials and equipment available in most teaching microbiology laboratories.
Figure 5 shows the results of a coaggregation assay. Tube one (on the left) is a pure culture showing no coaggregation, tube 2 (center tube) indicates a strong +4 reaction, while tube 3 (on the right) should be read as a +2 reaction.
These coaggregation exercises may be used with entire classes in courses such as general microbiology, allied health microbiology or environmental microbiology. Alternatively they may be used as a jumping off point for students doing independent research searching for coaggregation among environmental isolates from the mouth, fresh or marine water environments or soil. In this, students will be exploring a hot contemporary field of research in which investigators are attempting to determine just how general this phenomenon of coaggregation is in nature.
PREREQUISITES
Students should be able to define the term biofilm, describe the differences between biofilm (surface-attached) and planktonic (free-floating) bacteria, and describe why bacteria tend to grow on surfaces. The nature of adhesins should also be understood as well as the terms inter and intrageneric. Basic microbiological laboratory skills such as aseptic technique and transfer procedures should be mastered.
THE EXERCISES
The following exercises are designed to explore the interactions that exist between bacteria of the same genera (intrageneric coaggregations) and between bacteria of different genera (intergeneric coaggregations) in biofilms 3,4,5. The experiments are divided into three sections with the first section devoted to the exploration of coaggregations between four reference strains of oral bacteria. Some of the bacterial pairs coaggregate and some do not. Students may observe the clearing of the culture tube and quantify the strength and rate of the reaction by observing the degree of aggregation of the bacteria8.
The second section enables students to block coaggregation with lactose a competitive inhibitor of many lectin carbohydrate interactions.
The third section describes a method by which students may determine which of a pair of coaggregating bacteria possesses the lectin and which the cognate carbohydrate by using treatments (heat and protease enzymes) which denature the lectin protein.
ORAL BACTERIAL REFERENCE STRAINS
| Bacterium | Strain | Coaggregation Group |
|---|---|---|
| Actinomyces naeslundii | PK 35 | A |
| Streptococcus gordonii | ATCC 35105 | 1 |
| Streptococcus gordonii | ATCC 51656 | 6 |
| Streptococcus oralis | 34 | 3 |
| Streptococcus oralis | J22 | 4 |
Note: These strains are available from D. Clemans, Department of Biology, Eastern Michigan University, Ypsalanti MI., <daniel.clemans@emich.edu>
MATERIALS AND EQUIPMENT
| Per student or student group |
Item |
|---|---|
| 20 - 30 | 10 X 75mm or 12 X 75mm glass test tubes |
| 10 ml | Each of the reference strains in Table 1 |
| 4 ml | 300 mM lactose |
| 1 each | 200 μl, 20 μl, and 1000 μl pipettor with appropriate tips |
| Per Lab | Item |
|---|---|
| 1 | Refrigerated centrifuge |
| 1 | Spectrophtometer |
| 1 | 37° C Incubator |
| 1 | 85° waterbath or dry heat block |
INSTRUCTIONAL PROCEDURES
Prior to the laboratory period
1. Cultivation of Bacteria. All reference oral bacterial strains used in this study are listed in Table 1. Streptococci and actinomyces are cultured in Todd-Hewitt (TH) broth (BBL Microbiology Systems, Cockeysville, Md.) or Brain Heart Infusion (BHI) broth (BBL Microbiology Systems, Cockeysville, Md.) optimally at 37°C under anaerobic conditions with the GasPak system (BBL Microbiology Systems, Cockeysville, Md.). Theses strains, however, do grow well when cultured aerobically at 37°C on TH or BHI media.
2. Preparation of Bacteria for Coaggregation Assay. Bacterial cells used for coaggregation assays are pelleted by centrifugation at 10,000 X g for 10 min at 4°C. The supernatant is discarded and the cells are resuspended in coaggregation buffer. This wash step is repeated twice more (total of 3 washes). Coaggregation buffer consists of (1 mM Tris [pH 8.0], 0.1 mM CaCl2, 0.1 mM MgCl2, 150 mM NaCl, and 0.02% NaN3).
Adjust the bacterial suspensions to an A600 of approximately 1.0 (~109 bacteria/ml).
The cell suspensions thus prepared, are stored at 4°C until the laboratory period6,7,8.
Safety Notes: If the suspensions are to be used within a few weeks of the time of preparation, the sodium azide may be omitted.
All of the reference strains used in these exercises are listed as BSL 2 organisms. Instructors should take this into consideration when electing to carry out this exercise.
During the laboratory period –
Section one, measuring auto and coaggregation.
- Each group of students is provided with:
- 10 ml of each reference bacterial strain listed in Table 1.
- 20-30 glass tubes for coaggregation assays.
- 1 - 200 μl and 1- 20μl pipettor per group with 1-2 boxes of sterile tips. 1- 1000 μl pipettor with a box of sterile tips.
- Each individual strain should be tested for autoaggregation by placing 400 μl of the bacterium in a test tube. The tube is vortex mixed for 5 seconds and then rocked back and forth while observing aggregation formation. Any degree of settling or clearing of the tube indicates autoaggregation and this number (from the assessment scale) should be subtracted from the value measured for coaggregation.
- Pairs of organisms are tested for coaggregation by mixing 200 μl of each strain in a test tube. The tube is vortex mixed (~5 seconds) and then rocked back and forth until aggregates form. The degree of coaggregation is assessed by comparing the results with Table 2 and Figure 6 (Appendix). Visually score the coaggregates using the following 4-point scale8
Note: Good lighting is essential for viewing the extent of coaggregation. We have found that the surface of an overhead projector works well, a Quebec colony counter with its magnifying lens is even better.
- Record coaggregation data in tabular form for your report.
| Coaggregation Score | Description |
|---|---|
| 0 | Evenly turbid suspension of bacteria |
| 1 | Finely dispersed clumps in a turbid background |
| 2 | Definite clumps of bacteria are easily seen but do not settle immediately and remain in a turbid background |
| 3 | Clumps settle immediately with a slight turbid background |
| 4 | Clumps settle immediately and the supernatant is completely clear |
Section Two – Reversal of coaggregation with Lactose
Each group of students is provided with:
- 8 ml of 300 mM lactose (Other sugars e.g. N acetyl-glucosamine or N acetyl-galactosamine may be provided as well6
- Many Lectin carbohydrate interactions may be reversed by competitive inhibition with sugars such as lactose.
- Add 100 μl of a 300 mM stock solution of lactose to the 400 μl coaggregation assays from one of the existing coaggregation assays carried out in Section 1. This produces a final concentration of 60 mM lactose.
- The tubes are vortex mixed for 5 seconds and once again rocked while observing for coaggregation. If desired, other sugars can be added instead to a final concentration of 15 mM6.
- An easy way to keep track of each coaggregation assay is to set up a simple table with each strain represented on each axis (see references 6 and 7 for examples). Indicate the coaggregation score with lactose as a superscript to the original score without lactose (e.g. 40, 31 )
Section Three – Assessment of the Nature of the Interactions Between Bacterial Strains
Each group of students is provided with:
- An 85°C water bath or dry-heat block.
- To determine which partner contains the protein adhesin, incubate 2 ml of each bacterium at 85°C for 30 min, and then perform coaggregation as directed above (section 1).
Notes to Teachers:
- Coaggregation assays should be performed using both heated and unheated cells of the partner strains.
- Those bacterial strains that lose the ability, after heat treatment, to coaggregate with their partners contain the protein adhesin.
- Those treated bacterial strains that still coaggregate with their partners contain the carbohydrate receptor (See the proposed combinations and interpretations in Table 3).
- Each bacterial strain may possess an adhesin, a receptor or both (Figures 2 and 6).
| Assay | Interpretation |
|---|---|
| A x B | As in Section 1 |
| AH x B | A lowering of the Coaggregation score implies that A possesses a Lectin. |
| A x BH | A lowering of the Coaggregation score implies that B possess a Lectin |
| AH x BH | Further lowering of the Coaggregation score beyond either of the above trials implies that both strains possess Lectins (Bimodal Coaggregation) |
ASSESSMENT / EVALUATION
Assessment may be made by the instructor through visual evaluation of each student's coaggregation tube results as compared with a list of the known reactions for the microorganism pairs used (see appendix).
FOLLOW-UP ACTIVITIES
1) Coaggregation has been studied most intensively in dental plaque and to a lesser extent in fresh water environments. The field of investigation is wide open to explore coaggregation in any other natural (or human influenced) ecosystem. Organisms isolated from soil, sink drains, shower curtains, and any other habitat are potential materials for investigations of coaggregation.
Safety Note: Instructors should caution students about the small but real risk of isolating potential pathogens from natural habitats.
2) Students may explore bacterial coaggregation occurring in laboratory teaching strains of streptococci and other bacteria. The coaggregation properties of these strains, especially streptococci, can be assessed using the procedure followed in Section I.
- Culture the bacterial strain in an appropriate growth medium and wash the bacterial cells in coaggregation buffer as indicated above. Resuspend the bacterial suspension to a final cell density of A600 1.0 (~109 bacterial cells/ml.
- Assess the coaggregation properties as in Sections 2 and 3. Determine which partners contain the protein adhesin, and which strains contain the carbohydrate receptor.
REFERENCES
- Gibbons, R. J. and M. Nygaard, 1970, Interbacterial Aggregation of Plaque Bacteria. Archives of Oral Biology 15:1397-1400.
- Kolenbrander, P.E., and R.J. Palmer Jr., 2004 Human Oral Bacterial Biofilms, In M. Ghannoum and G.A.O’Toole, Microbial Biofilms. ASM Press, Washington D.C.
- Rickard, A.H. P. Gilbert, N.J. High, P.E. Kolenbrander, and P.S. Handley. 2003. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends in Microbiology 11:94-100.
- Kolenbrander, P.E., R.N. Andersen, D.S. Blehert, P.G. Egland, J.S. Foster, and R.J. Palmer Jr. 2002. Communication among oral bacteria. Microbiol. Molec. Biol. Rev. 66:486-505.
- Kolenbrander, P.E. 1989. Surface recognition among oral bacteria: multigeneric coaggregation and their mediators. Crit. Rev. Microbiol. 17:137-159.
- Kolenbrander, P.E., R.N. Andersen, and L.V. Moore. 1990. Intrageneric coaggregation among strains of human oral bacteria: potential role in primary colonization of the tooth surface. Appl. Environ. Microbiol. 56:3890-3894.
- Clemans, D.L., and P.E. Kolenbrander. 1995. Isolation and characterization of coaggregation-defective (Cog-) mutants of Streptococcus gordonii DL1 (Challis). J. Industrial Microbiol. 15:193-197.
- Kolenbrander, P.E. 1988. Intergeneric coaggregation among human oral bacteria and ecology of dental plaque. Ann. Rev. Microbiol. 42:627-656.
Appendix
Coaggregation of Actinomyces naeslundii PK35 with selected reference strains of streptococci
| S. gordonii ATCC35105 (1) |
S. oralis 34 (3) | S. oralis J22 (4) | S. gordonii ATCC51656 (6) |
A. naeslundii PK35 (A) |
|
|---|---|---|---|---|---|
| S. gordonii ATCC35105 (1) | 30 | 0 | 33 | ||
| S. oralis 34 (3) | 30 | 30 | |||
| S. oralis J22 (4) | 43 | ||||
| S. gordonii ATCC51656 (6) | |||||
| A. naeslundii PK35 (A) |
aCoaggregation scores are given in two parts: the first score is that given after mixing the two strains together, and the second score (superscript) is that given after adding lactose to a final concentration of 60 mM.
Streptococcal-streptococcal coaggregations: Kolenbrander, P.E., R.N. Andersen, and L.V.H. Moore. 1990.Intrageneric coaggregation among strains of human oral bacteria: potential role in primary colonization of the tooth surface. Infect. Immun. 56:3890-3894. Clemans, D.L., and P.E. Kolenbrander. 1995. Isolation and characterization of coaggregation-defective (Cog-) mutants of Streptococcus gordonii DL1 (Challis). J. Indust. Microbiol. 15:193-197.
Streptococcus-actinomyces coaggregations: Cisar, J.O., P.E. Kolenbrander, and F.C. McIntire. 1979. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect. Immun. 24:742-752. Egland, P.G., L.D. Du, and P.E. Kolenbrander. 2001. Identification of independent Streptococcus gordonii SspA and SspB functions in coaggregation with Actinomyces naeslundii. Infect. Immun. 69:7512-7516.
Figure 6 shows a diagramatic representation of coaggregations between reference oral bacteria used in this exercise. Each interaction is depicted as a complementary set of symbols. Symbols with stems (adhesions) represent heat-inactivated components on the surface of the respective bacterial strains. The complementary rectangular or triangular symbols, without stems (receptors), represent components that are resistant to heat treatment and are thought to be receptors composed of carbohydrates.
References to the origin of the strains used in this exercise:
Lactose-sensitive coaggregations between streptococci.
Clemans, D.L., and P.E. Kolenbrander. 1995. Isolation and characterization of coaggregation-defective (Cog-) mutants of Streptococcus gordonii DL1 (Challis). J. Indust. Microbiol. 15:193-197.
Kolenbrander, P.E., R.N. Andersen, and L.V.H. Moore. 1990. Intrageneric coaggregation among strains of human oral bacteria: potential role in primary colonization of the tooth surface. Appl. Environ. Microbiol. 56:3890-3894.
Lactose-sensitive and lactose-insensitive coaggregations between streptococci and actinomyces.
Egland, P.G., L.D. DU, and P.E. Kolenbrander. 2001. Identification of Independent Streptococcus gordonii SspA and SspB Functions in Coaggregation with Actinomyces naeslundii. Infect. Immun. 69:7512-7516.
Kolenbrander, P.E. 1988. Intergeneric coaggregation among human oral bacteria and ecology of dental plaque. Ann. Rev. Microbiol. 42:627-656.
Kolenbrander, P.E. R.N. Andersen, D.S. Blehert, P.G. Egland,
J.S. Foster, and R.J. Palmer Jr. 2002. Communication among oral bacteria. Microbiol. Molec. Biol. Rev. 66:486
